As a Senior Chief AI Engineer at Samsung R&D, I’ve spent the last few years working at the intersection of machine learning and human health. From improving smartphone vision systems to decoding heartbeats, I’ve seen Convolutional Neural Networks (CNNs) evolve into powerful tools beyond just image recognition.
In this blog, I’ll unravel the “why” behind CNNs, show you how they preserve meaningful structure in data, and share how we’re adapting them to save lives—one heartbeat at a time.
The CNN Advantage: Preserving Spatial Relationships
Imagine trying to understand a sentence after randomly shuffling its words—that’s what traditional neural networks do to images. Flattening a 256×256 MRI scan into a 1D vector throws away the spatial relationships that matter most.
# Problem: Flattening a 256×256 MRI scan
flattened = image.reshape(1, 256*256*3) # 196,608 features!
# Loses all spatial context between pixels
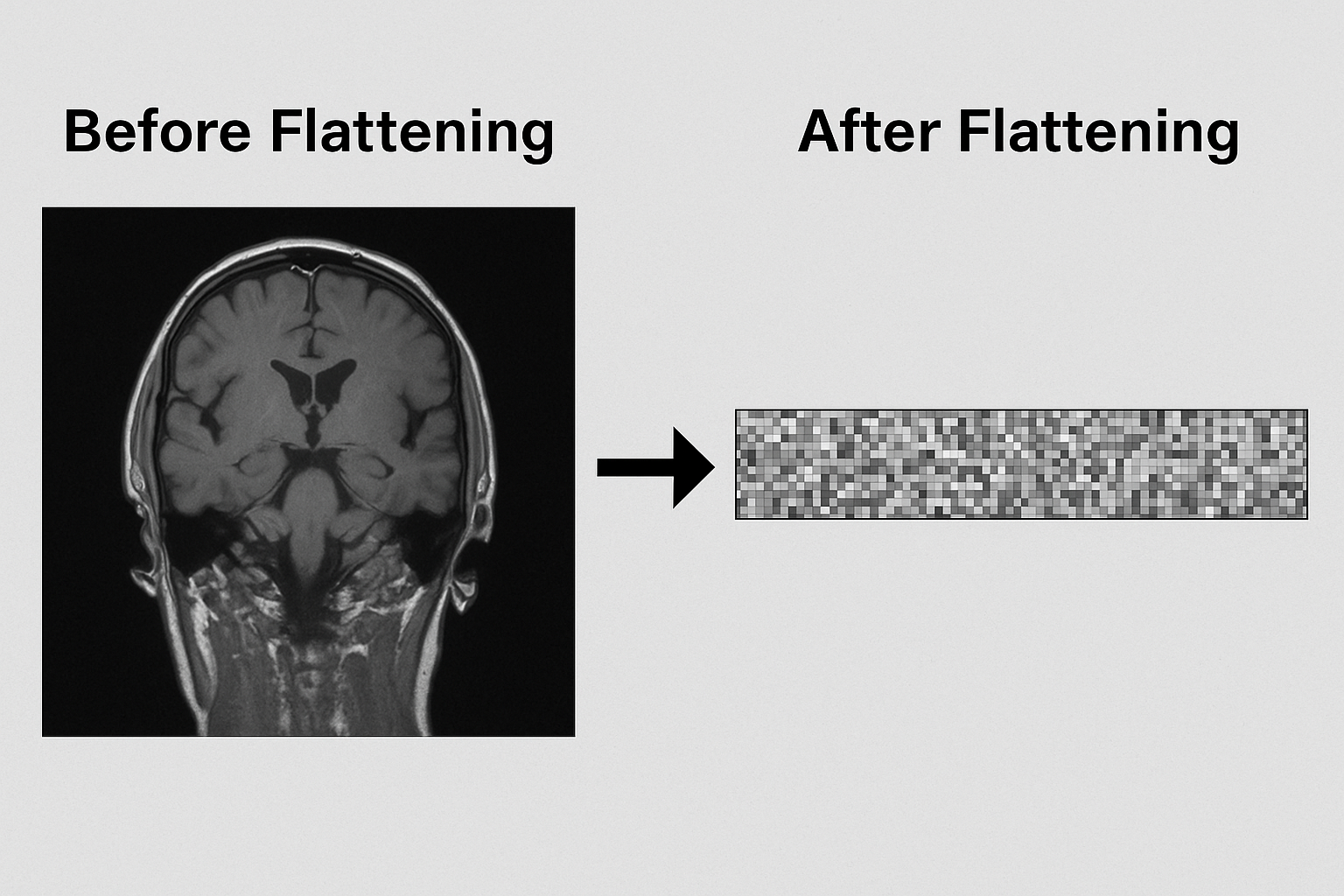
Flattening destroys local relationships
That’s where CNNs shine:
- Local connectivity: Filters look at small patches (receptive fields)
- Parameter sharing: The same kernel slides across the image
- Hierarchical learning: From simple edges to complex organs
Core CNN Operations (2D Example)
Core CNN Operations (2D Example)
Let's break down the key operations that make CNNs so powerful for spatial data:
1. Convolution: The Pattern Detector
# Typical 2D convolution in PyTorch/Keras
conv_layer = Conv2D(filters=32,
kernel_size=(3,3),
strides=1,
padding='same')
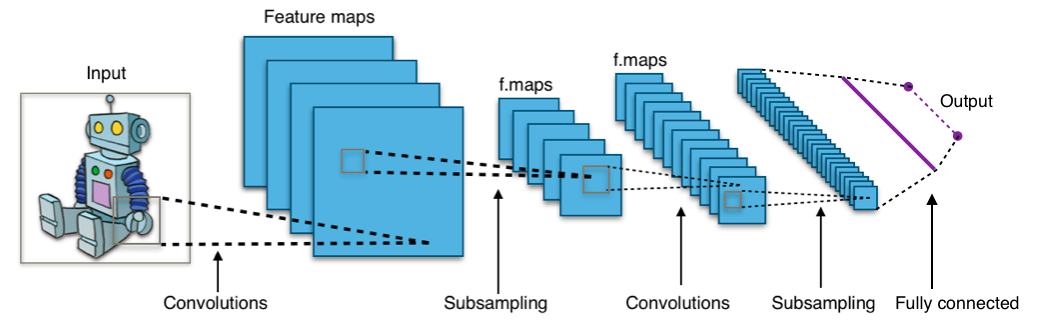
Three-layer CNN architecture showing feature map evolution
- What happens: A 3×3 kernel slides across the image, computing dot products at each position
- Why it matters: Learns local features like edges, textures, or medical anomalies
- Key parameters:
- Filters: Number of feature detectors (32 here)
- Kernel size: Receptive field (3×3 is standard)
- Stride: How many pixels to shift (1=high resolution)
2. Pooling: The Information Compressor
# Max pooling reduces spatial dimensions
pool_layer = MaxPool2D(pool_size=(2,2),
strides=2)
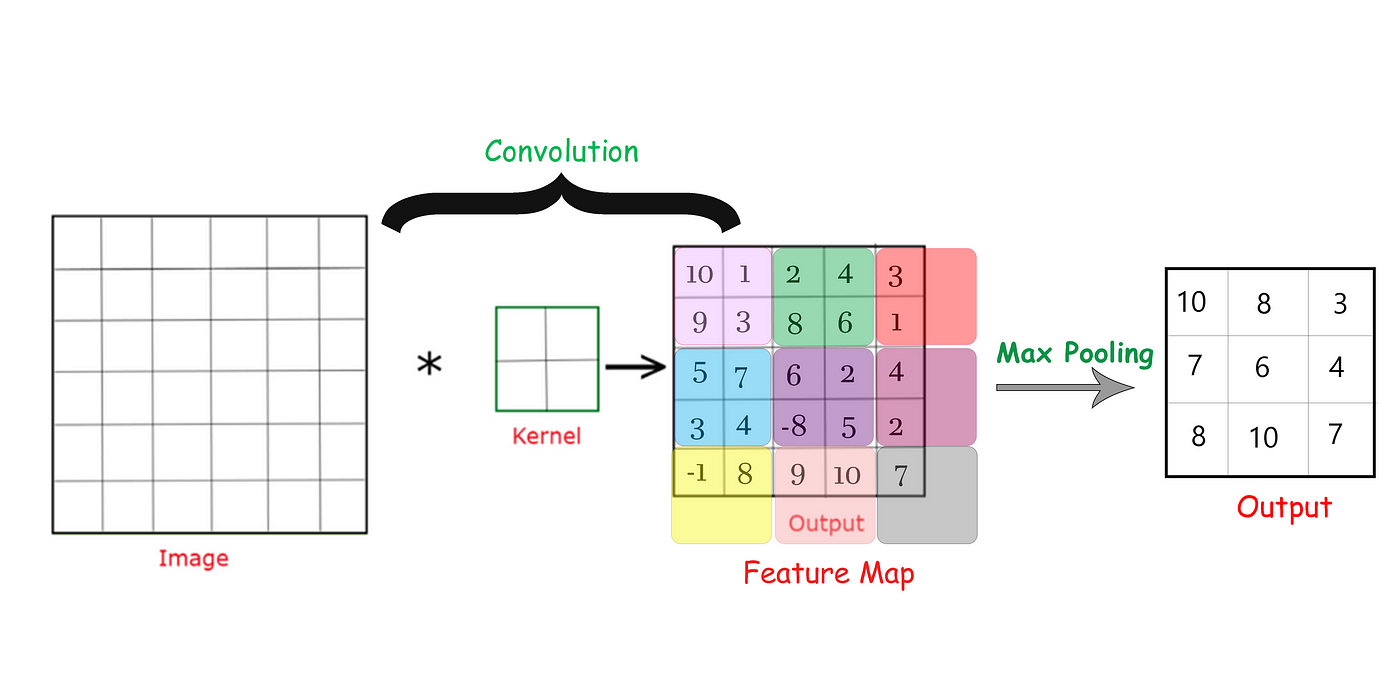
Max pooling operation preserving strongest activations
- What happens: Downsamples feature maps by taking maximum values in 2×2 windows
- Why it matters:
- Makes network invariant to small translations
- Reduces computational load
- Expands receptive field without adding parameters
- Medical imaging tip: Sometimes use average pooling for smoother feature maps
3. Activation: The Non-Linear Transformer
# ReLU activation (standard for CNNs)
activation = ReLU()
# or LeakyReLU(alpha=0.1) for medical data
- Mathematically: max(0, x) - zeros out negative activations
- Why ReLU?:
- Mitigates vanishing gradients
- Computationally efficient
- Encourages sparse activations (helpful for localized medical features)
- Medical variant: LeakyReLU often works better for ECG/EEG data
Putting It All Together
A typical CNN block for medical imaging:
Sequential(
Conv2D(64, (3,3), padding='same'),
BatchNormalization(),
ReLU(),
MaxPool2D((2,2)),
Dropout(0.2)
)
💡
Pro Tip: For medical images, we often use smaller strides (1 instead of 2)
in early layers to preserve fine diagnostic details that might indicate early-stage pathology.
From Pixels to Pulses: CNNs for 1D Signals
What if I told you the same magic behind facial recognition also helps decode heart rhythms? While CNNs were born in the image world, their DNA works just as well for 1D signals like ECGs.
Think about it: An ECG is like a one-dimensional image. Local waveform patterns like P-waves and QRS complexes hold crucial information—exactly the kind of features CNNs are good at extracting.
1D Convolution for ECG Analysis
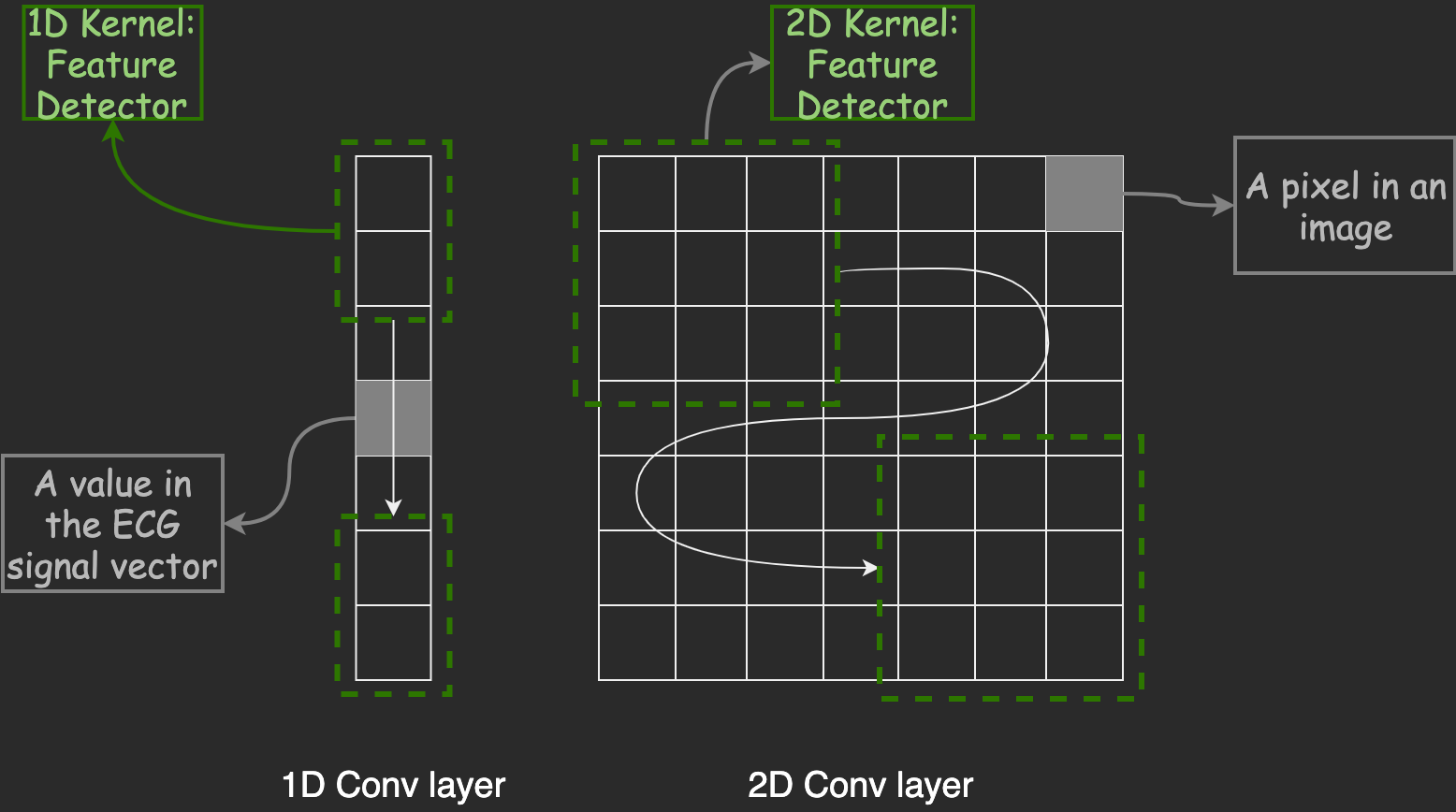
CNN kernels sliding over ECG signals to detect QRS complexes—just like detecting edges in images
CNN kernels sliding over ECG signals to detect QRS complexes—just like detecting edges in images
# 1D vs 2D convolution comparison
# 2D (images) # 1D (ECG)
Conv2D(32, (3,3)) Conv1D(32, 3)
Input: (256,256,3) Input: (1000,1) # 1000 samples
Case Study: Cleaning Noisy ECGs with CNNs + Attention
In one of my recent research projects, we developed a hybrid deep learning model called the Attention-Based Convolutional Denoising Autoencoder (ACDAE). Our mission: Clean up highly noisy two-lead ECG signals and accurately detect arrhythmias, even under extreme real-world noise conditions.
The ACDAE architecture blends the strengths of convolutional autoencoders and attention mechanisms:
- Encoder: 1D convolutional layers extract compressed, noise-resilient features from raw ECG signals.
- Skip Connections: Direct links between encoder and decoder layers help recover fine-grained cardiac signal details lost during downsampling.
- Decoder: 1D transposed convolutional layers reconstruct the clean ECG signals from compressed features.
- Attention (ECA Module): An efficient channel attention mechanism dynamically focuses on the most diagnostically relevant parts of the signal during reconstruction and classification.
1. Noise Robustness
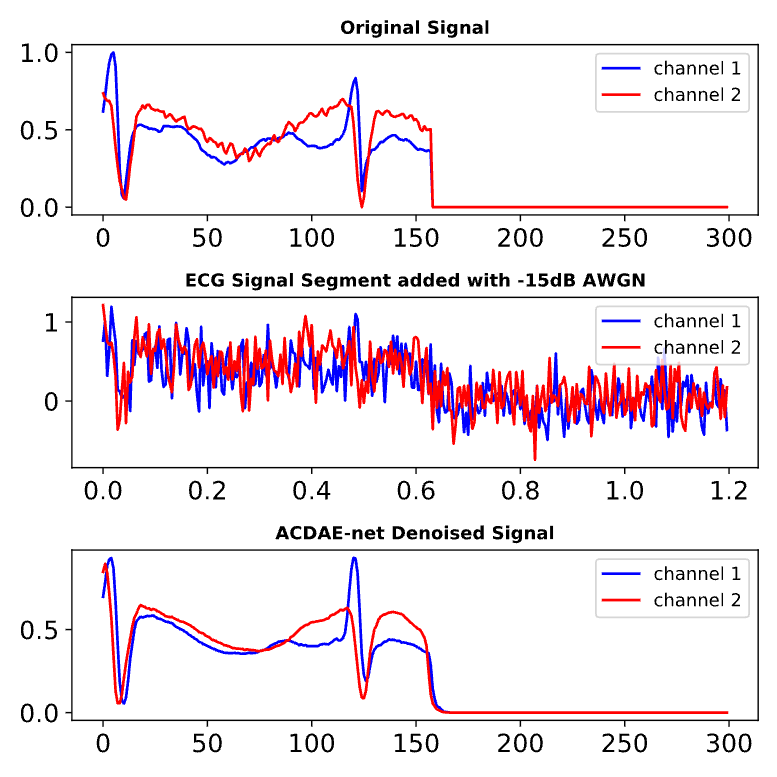
The denoising autoencoder filters out movement artifacts and high-frequency noise, ensuring that clinical features (like P and T waves) remain intact for accurate diagnosis.
2. Attention Mechanism
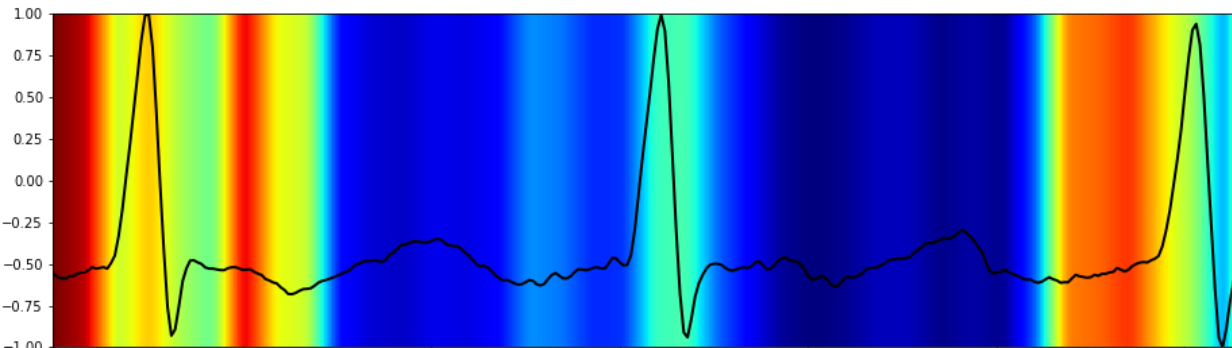
Attention gates learn to prioritize signal segments with clinical significance—such as an ST-segment elevation—enhancing interpretability and model trust.
We evaluated ACDAE across four major ECG databases and under varying noise levels from -20 dB to +20 dB SNR. The model achieved impressive results:
- 19.07 dB average SNR improvement (even under extreme noise).
- 98.88% arrhythmia classification accuracy with attention-enhanced features.
- Lightweight enough for potential deployment on wearable healthcare devices.
Under the Hood:
- Encoder: Stacked 1D Conv layers with Leaky ReLU activations
- Attention Bottleneck: Efficient Channel Attention (ECA) modules to highlight important cardiac features
- Decoder: 1D Transposed Conv layers reconstruct the clean signal
- Classifier: A compact fully-connected path for atrial fibrillation (AF) detection
Read the full IEEE paper
Practical Tips: Images vs Signals
Medical signals behave differently from natural images. Here's a cheat sheet I always keep in mind:
| Factor |
Natural Images |
Medical Signals (e.g. ECG) |
| Augmentation |
Flip, rotate freely |
Use time-warping, scaling only |
| Kernel Size |
Typically 3×3 |
Depends on waveform duration (e.g., QRS ≈ 120ms) |
Takeaway: The CNN Mindset
Whether I’m analyzing X-rays or heartbeats, my approach remains the same:
- Look locally: Slide filters to extract small patterns
- Go deep: Stack layers to build abstraction
- Tune wisely: Adapt kernel shapes to the data’s domain
CNNs are no longer just for selfies or cat videos—they’re helping diagnose heart disease, detect tumors, and even enhance driver safety. And as we adapt them to new domains, their impact only grows.
Thanks for reading! Feel free to connect if you're working on AI in healthcare—I’d love to learn from your journey too.






