From Concept to Implementation: A Comprehensive Guide
In this deep dive, we'll build a complete Retrieval-Augmented Generation system using SQLite as our knowledge base. I'll explain each component in detail, ensuring you understand not just how it works, but why it works.
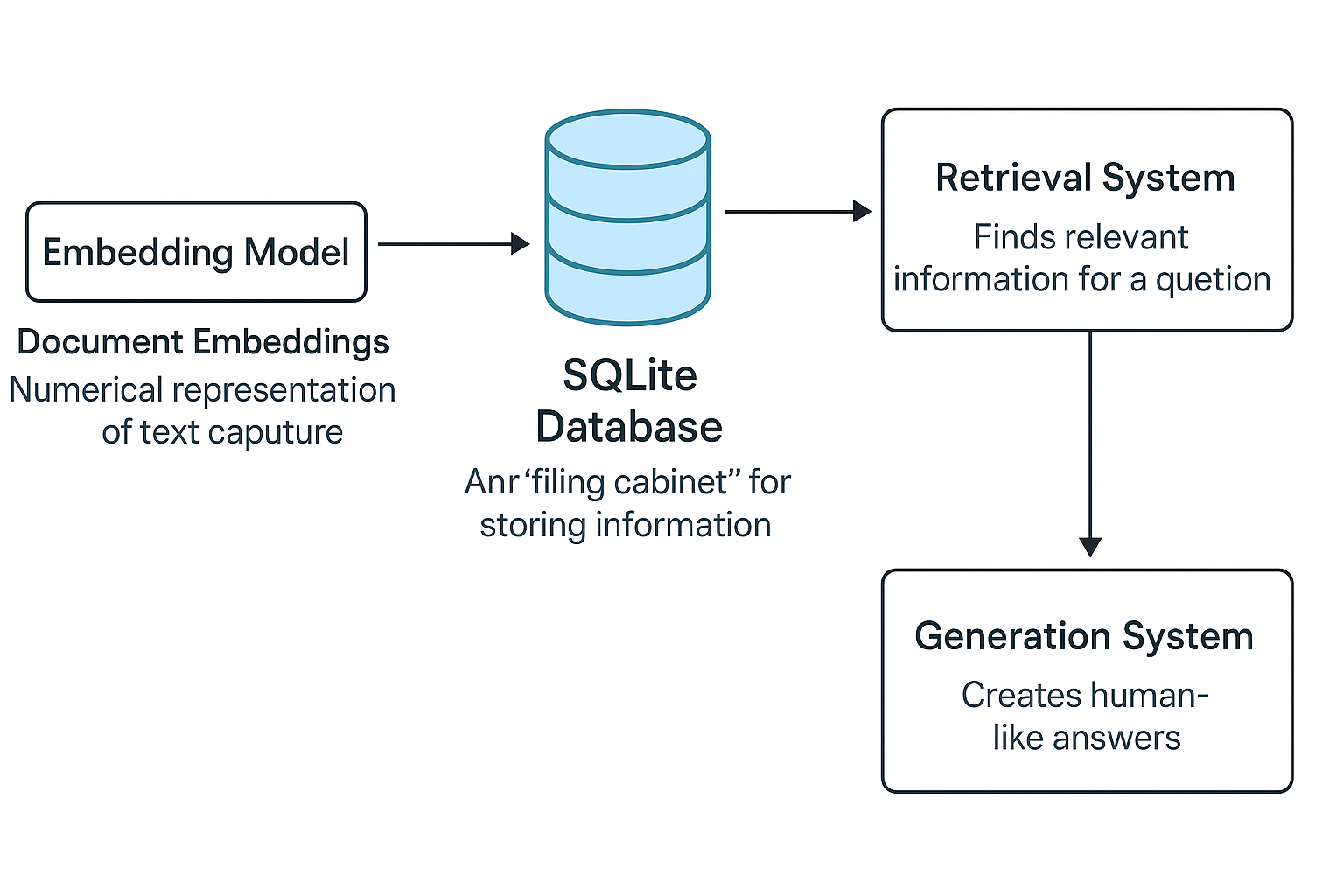
1. Database Setup: Creating Our Knowledge Foundation
SQLite serves as the backbone of our RAG system - it's where we'll store all our knowledge documents and their vector embeddings.
Why SQLite?
- Serverless Architecture: Unlike MySQL or PostgreSQL, SQLite doesn't require a separate server process
- Single-File Storage: The entire database is stored in one cross-platform file
- Full SQL Support: We get all the power of SQL queries without setup complexity
The Documents Table Structure
We create a table with four critical columns:
CREATE TABLE IF NOT EXISTS documents (
id INTEGER PRIMARY KEY, -- Unique identifier for each document
title TEXT, -- Short descriptive title
content TEXT, -- Full text content
embedding BLOB -- Vector representation stored as binary
)The BLOB (Binary Large Object) type is perfect for storing our vector embeddings which are essentially arrays of floating-point numbers.
2. Populating the Knowledge Base
Our sample dataset focuses on Python programming concepts:
documents = [
("Python Lists", "Python lists are ordered, mutable collections..."),
("Dictionaries", "Dictionaries store key-value pairs..."),
("Functions", "Functions are reusable code blocks...")
]Insertion Process Explained
For each document, we:
- Execute an INSERT statement with the title and content
- Initially leave the embedding NULL (we'll add it later)
- Commit the transaction to make changes permanent
for title, content in documents:
cursor.execute('''
INSERT INTO documents (title, content)
VALUES (?, ?) -- Parameterized query prevents SQL injection
''', (title, content))
conn.commit() # Save changes to disk3. The Magic of Text Embeddings
Embeddings transform text into numerical vectors that capture semantic meaning.
Choosing the Right Model
We use all-MiniLM-L6-v2 because:
- It's small (80MB) but effective
- Generates 384-dimensional vectors
- Balances speed and accuracy well
from sentence_transformers import SentenceTransformer
model = SentenceTransformer('all-MiniLM-L6-v2')
text = "Python lists are ordered collections..."
embedding = model.encode(text) # Returns numpy array of shape (384,)Embedding Properties
These vectors have special properties:
- Similar texts have similar vectors
- We can measure similarity using cosine distance
- The model understands context (e.g., "bank" as financial vs river)
4. Storing Embeddings Efficiently
We need to store these numpy arrays in SQLite:
# Convert numpy array to bytes for storage
embedding_bytes = embedding.tobytes()
# Update the database
cursor.execute('''
UPDATE documents
SET embedding = ?
WHERE id = ?
''', (embedding_bytes, doc_id))Why BLOB Storage?
- SQLite doesn't natively support array types
- BLOBs efficiently store binary data
- We'll convert back to numpy when retrieving
5. The Retrieval Process
When a user asks a question, we:
- Embed the question
- Compare against all document embeddings
- Return the most relevant matches
Understanding Cosine Similarity
This measures how similar two vectors are:
import numpy as np
def cosine_similarity(a, b):
return np.dot(a, b) / (np.linalg.norm(a) * np.linalg.norm(b))Returns:
- 1 = identical meaning
- 0 = no relationship
- -1 = opposite meaning
Implementing the Retrieval
We create a custom function to compare embeddings:
def retrieve(query, top_k=3):
query_embedding = model.encode(query)
# Get all documents
cursor.execute("SELECT id, title, content, embedding FROM documents")
results = []
for doc_id, title, content, embedding_bytes in cursor.fetchall():
doc_embedding = np.frombuffer(embedding_bytes, dtype=np.float32)
similarity = cosine_similarity(query_embedding, doc_embedding)
results.append((title, content, similarity))
# Sort by similarity (descending)
results.sort(key=lambda x: x[2], reverse=True)
return results[:top_k]6. Generating Responses
With relevant documents retrieved, we generate an answer:
def generate_response(query, relevant_docs):
# Prepare context
context = "\n\n".join(
f"Document {i+1} ({sim:.2f}): {content[:200]}..."
for i, (title, content, sim) in enumerate(relevant_docs)
)
# In production, we'd use an LLM here
return f"""Question: {query}
Relevant Context:
{context}
Summary Answer: {relevant_docs[0][1][:300]}..."""Production Considerations
In a real system, you would:
- Replace the simple concatenation with an actual LLM call
- Implement proper prompt engineering
- Add citation of sources
Complete System Architecture
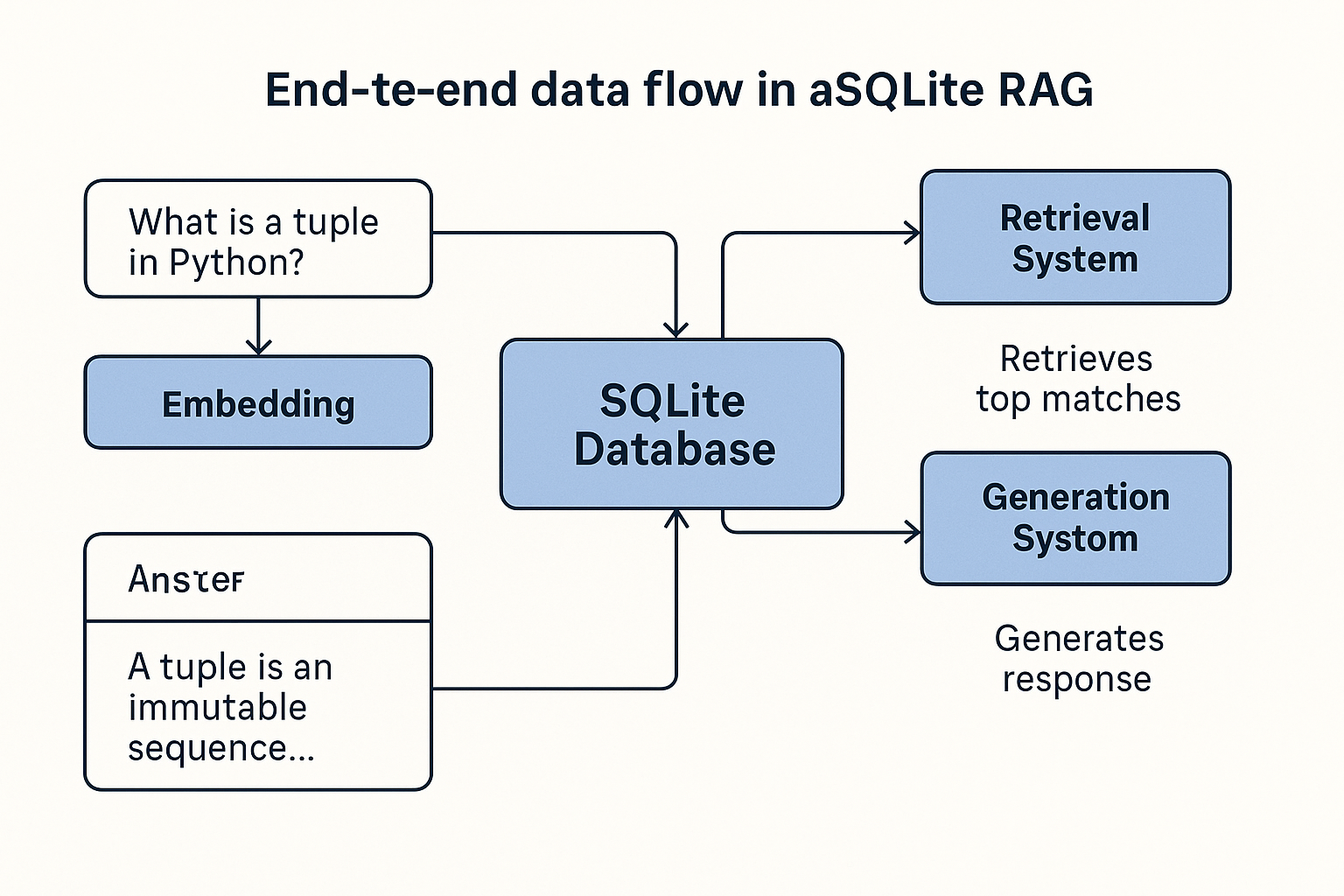
Component Interaction
- User submits a question
- System converts question to embedding
- Compares against stored document embeddings
- Retrieves top matches from SQLite
- Generates response using context
- Returns final answer to user
Scaling Considerations
For production systems, consider:
Performance Optimizations
- Indexing: Add indexes on embedding columns
- Chunking: Split large documents into smaller passages
- Caching: Store frequent queries
- Hybrid Search: Combine with keyword search
When to Upgrade
SQLite works great for:
- Prototyping and small datasets (<10,000 documents)
- Applications with low query volume
- Systems where simplicity is prioritized
Consider specialized vector databases (Chroma, Weaviate) when:
- Your dataset grows beyond 50,000 documents
- You need advanced similarity search features
- Your application requires high query throughput